The team of Department of Genetics, Institute of Biology and Ecology, Faculty of Science, P. J. Šafárik University in Košice under the leadership of prof. RNDr. Eva Čellárová, DrSc. has been engaged in research of natural compounds with significant pharmacological and therapeutic potential for many years. These are mainly bioactive anthraquinones, primarily hypericin. Some, but not all representatives of the genus Hypericum are the only producers of hypericin in the plant kingdom. The availability of hypericin and other bioactive substances on the world market is ensured essentially by Hypericum perforatum.
In addition to plants representing predominantly by the Hypericum species, the anthraquinones including hypericin, and its putative intermediates emodin, emodin anthrone, skyrin and others are widespread in fungi including fungal plant-borne endophytes. Biosynthetic core genes and genes coding for regulatory elements are clustered in the fungal genomes and reveal fundamental aspects of their potential genetic and regulatory function at the genomic level. Understanding of genetic and epigenetic background of the anthraquinone biosynthetic gene clusters and way of their activation in fungal endophytes would help not only understand their pathways in plants, especially Hypericum spp. as the essential producers of hypericin in the plant kingdom, but also favour them as promising heterologous/homologous system for prospective biotechnological production of hypericin and other desirable anthraquinones.
The team of Department of Genetics, Institute of Biology and Ecology, Faculty of Science, P. J. Šafárik University in Košice under the leadership of prof. RNDr. Eva Čellárová, DrSc. has been engaged in research of natural compounds with significant pharmacological and therapeutic potential for many years. These are mainly bioactive anthraquinones, primarily hypericin. Some, but not all representatives of the genus Hypericum are the only producers of hypericin in the plant kingdom. The availability of hypericin and other bioactive substances on the world market is ensured essentially by Hypericum perforatum.
In addition to plants representing predominantly by the Hypericum species, the anthraquinones including hypericin, and its putative intermediates emodin, emodin anthrone, skyrin and others are widespread in fungi including fungal plant-borne endophytes. Biosynthetic core genes and genes coding for regulatory elements are clustered in the fungal genomes and reveal fundamental aspects of their potential genetic and regulatory function at the genomic level. Understanding of genetic and epigenetic background of the anthraquinone biosynthetic gene clusters and way of their activation in fungal endophytes would help not only understand their pathways in plants, especially Hypericum spp. as the essential producers of hypericin in the plant kingdom, but also favour them as promising heterologous/homologous system for prospective biotechnological production of hypericin and other desirable anthraquinones.
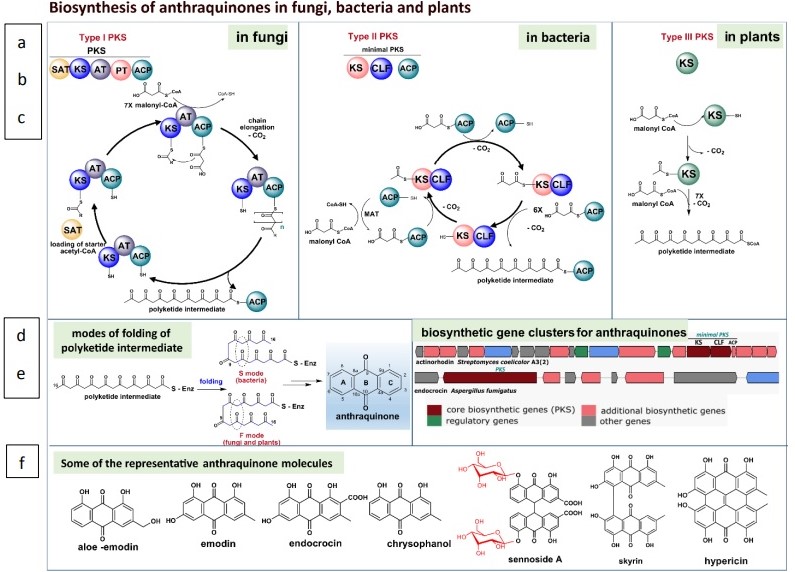
Biosynthesis of anthraquinones in fungi, bacteria, and plants by (a) Type I, (b) Type II and (c) Type III PKS (polyketide synthase), respectively. PKSs condenses starter acyl CoAs (typically acety-CoA) with seven malonyl-CoAs to form an ACP bound linear polyketide chain. (d) The polyketide chain undergoes two different modes of folding, S-Mode (in bacteria) and F-Mode (in fungi and plants). The genes in bacterial (actinorhodin) and fungal (endocrocin) biosynthetic gene clusters coding for some anthraquinones (adopted form antiSMASH database) (e). Examples of anthraquinones and bisanthraquinones (f).
SAT- starter unit ACP transacylase
AT- acyltransferase
KS – ketosynthase
CLF – chain length factor
PT- product template
ACP – acyl carrier protein
Different groups of anthraquinones synthesized by diverse and phylogenetically distant groups of organisms raise the question: what genes and enzymes of the biosynthetic pathway are evolutionary conserved orthologues and what changes in the genomes over time led to a wide spectrum of anthraquinone derivatives? An increasing evidence on genome organization reveals that the polyketide biosynthetic genes in bacteria and fungi are clustered, but the data on the organization of the polyketide genes in plant genomes is scarce. Another important aspect is the communication between the symbiotic organisms and their plant hosts and its impact on the biosynthetic capability. Is it necessary, for prospective production purposes, to elucidate the entire pathway or just to focus on the key reaction(s) necessary for application of the synthetic biology approach?
More information available in the recently published paper of N.K. Mund (postdoctoral fellow at the Department of Genetics) and E. Čellárová:
Mund, NK. & Čellárová, E.: Recent Advances in the identification of biosynthetic genes and gene cluste
rs of the polyketide-derived pathways for anthraquinone biosynthesis and biotechnological applications. Biotechnological Advances 63 (2023) 108104