Džupponová V, Žoldák G. Aggregation mechanism and branched 3D morphologies of pathological human light chain proteins under reducing conditions. Colloids Surf B Biointerfaces. 2023 Jan;221:112983. doi: 10.1016/j.colsurfb.2022.112983.
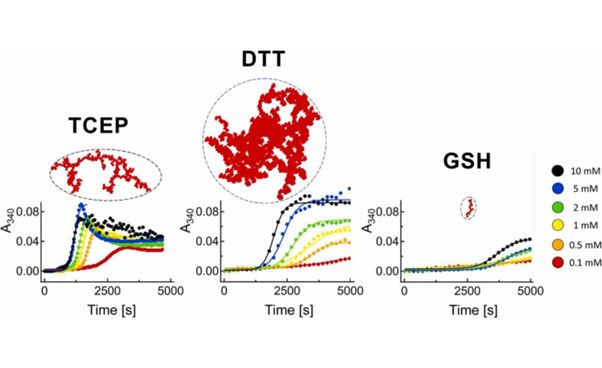
Multiple myeloma is a hematologic malignancy characterized by a clonal proliferation of plasma cells producing a monoclonal immunoglobulin light chain. Produced monoclonal light chains are then secreted in the blood serum and called free light chains, which circulate and can start forming extracellular sediments at different locations, including vital organs. Here, we examined the aggregation mechanism and structures of the pathological human multiple myeloma light chain aggregates (hLC) after disrupting stabilizing disulfide bonds by various reducing agents. The aggregation kinetics were measured in the presence of three commonly used disulfide reducers (TCEP, DTT and glutathione), and the resulting aggregates were visualized by the combination of light and confocal/super-resolution STED microscopy. We find that aggregation kinetics can be described by two apparent macroscopic rate constants of the Finke-Watzky model related to the nucleation and the growth process. Surprisingly, the growth rate constants decreased at higher protein concentrations, which we interpret as the involvement of an aggregation active monomer particle that is successively depleted at high concentrations due to shifts in a monomer/dimer equilibrium. Seeding experiments demonstrated the specificity of the aggregates; only certain seeds accelerated the aggregation, while others eventually slowed down the aggregation. Three-dimensional visualization of the overall structures of the final aggregates at submicrometer resolution showed variable, reducer-specific branched morphologies with non-trivial fractal dimensions. Thus, the disruption of the stabilizing disulfide bonds in hLC leads to specific large, branched aggregates formed by the monomer-addition mechanism.




